Data Imputation using Autoencoders | What to do when data is missing? (Part II)
— Data Imputation, Neural Networks, Autoencoders, Python — 4 min read
Share
Mushrooms, anyone? What if you have lots of data on mushrooms, yet some of it is missing? One important question you might want to answer is whether or not a particular specimen is edible or poisonous. Of course, your understanding of what a poisonous mushroom is might be quite different (hi to all from Netherlands), but I digress.
The dataset of interest will be (you guessed it) all about mushrooms. It describes physical characteristics of 8124 mushroom instances. The number of variables is 23, all of which are categorical. More information about the dataset can be found here.
1. Autoencoders
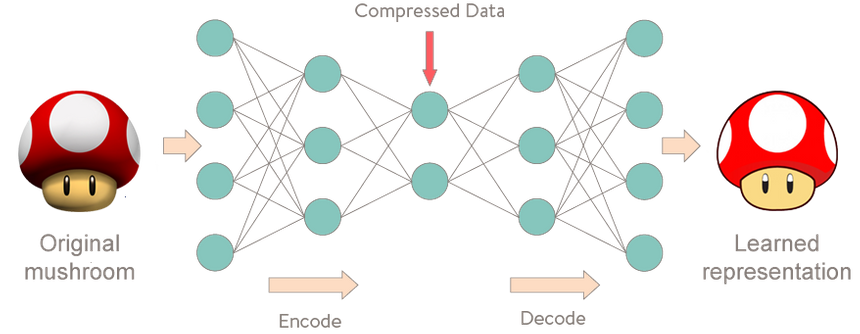
Strangely enough, an autoencoder is a model that given input data tries to predict it. It is used for unsupervised learning (That might not be entirely correct). Puzzling? First time I heard the concept I thought it must be a misunderstanding on my part. It wasn’t.
More specifically, let’s take a look at Autoencoder Neural Networks. This autoencoder tries to learn to approximate the following identity function:
fW,b(x)≈xWhile trying to do just that might sound trivial at first, it is important to note that we want to learn a compressed representation of the data, thus find structure. This can be done by limiting the number of hidden units in the model. Those kind of autoencoders are called undercomplete.
1.1 Choosing loss function
In order to learn something meaningful, autoencoders should try to minimize some function, called reconstruction error. The traditional squared error is often used:
L(x,x′)=∣∣x−x′∣∣22.1 Encoding the data
Our dataset contains categorical variables exclusively. We will use a standard approach for such cases - one-hot encoding. Furthermore, we have to handle cells with missing values. We will create a missing mask vector and append it to our one-hot encoded values. Missing values will be filled with some constant.
Let’s take a look at this sample data:
1[2 ['blue stem', 'large cap'],3 [None, 'large cap'],4 ['green stem', None]5]Our encoded data looks like this:
1[2 [1, 0, 0, 1, 0, 0, 0, 0, 0, 0], # no missing values3 [0, 0, 1, 1, 0, 1, 1, 1, 0, 0], # missing value in 1st variable4 [0, 1, 0, 0, 1, 0, 0, 0, 1, 1] # missing value in 2nd variable5]2.2 Reconstruction error
Our reconstruction error will be the mean squared error which hopefully will work for categorical data. Since we are using Keras, our function must adhere to some rules.
1def make_reconstruction_loss(n_features):23 def reconstruction_loss(input_and_mask, y_pred):4 X_values = input_and_mask[:, :n_features]5 X_values.name = "$X_values"67 missing_mask = input_and_mask[:, n_features:]8 missing_mask.name = "$missing_mask"9 observed_mask = 1 - missing_mask10 observed_mask.name = "$observed_mask"1112 X_values_observed = X_values * observed_mask13 X_values_observed.name = "$X_values_observed"1415 pred_observed = y_pred * observed_mask16 pred_observed.name = "$y_pred_observed"1718 return mse(y_true=X_values_observed, y_pred=pred_observed)19 return reconstruction_lossAdditionally, we will use slightly modified mean squared error for assessing our progress during training. The function takes into account the mask of the input data:
1def masked_mae(X_true, X_pred, mask):2 masked_diff = X_true[mask] - X_pred[mask]3 return np.mean(np.abs(masked_diff))2.3 Getting everything just right
Of course, a couple of imports are needed to make everything work. Make sure you have the proper libraries installed (all available via pip install).
1import random2import pandas as pd3import numpy as np4import matplotlib.pylab as plt5import seaborn as sns6from keras.utils import np_utils7from sklearn.preprocessing import LabelEncoder89from keras.objectives import mse10from keras.models import Sequential11from keras.layers.core import Dropout, Dense12from keras.regularizers import l1l21314from collections import defaultdict1%matplotlib inline2.4 Creating the model (the fun part)
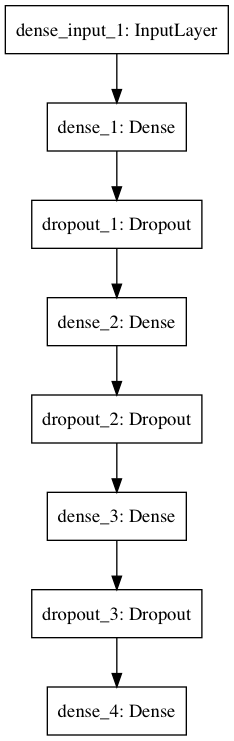
The following implementation is heavily based on the one provided by fancyimpute. Our NN has 3 hidden layers (with ReLU activation functions) with dropout probability set to 0.5. You can also choose two regularizers coefficients - the sum of the weights (L1) and sum the squares of the weights (L2). These are 0. The activation function for the output layer is sigmoid. It appears to work better than linear for our use case.
Here is the code for it, don’t be afraid to fiddle with it:
1class Autoencoder:23 def __init__(self, data,4 recurrent_weight=0.5,5 optimizer="adam",6 dropout_probability=0.5,7 hidden_activation="relu",8 output_activation="sigmoid",9 init="glorot_normal",10 l1_penalty=0,11 l2_penalty=0):12 self.data = data.copy()13 self.recurrent_weight = recurrent_weight14 self.optimizer = optimizer15 self.dropout_probability = dropout_probability16 self.hidden_activation = hidden_activation17 self.output_activation = output_activation18 self.init = init19 self.l1_penalty = l1_penalty20 self.l2_penalty = l2_penalty2122 def _get_hidden_layer_sizes(self):23 n_dims = self.data.shape[1]24 return [25 min(2000, 8 * n_dims),26 min(500, 2 * n_dims),27 int(np.ceil(0.5 * n_dims)),28 ]2930 def _create_model(self):3132 hidden_layer_sizes = self._get_hidden_layer_sizes()33 first_layer_size = hidden_layer_sizes[0]34 n_dims = self.data.shape[1]3536 model = Sequential()3738 model.add(Dense(39 first_layer_size,40 input_dim= 2 * n_dims,41 activation=self.hidden_activation,42 W_regularizer=l1l2(self.l1_penalty, self.l2_penalty),43 init=self.init))44 model.add(Dropout(self.dropout_probability))4546 for layer_size in hidden_layer_sizes[1:]:47 model.add(Dense(48 layer_size,49 activation=self.hidden_activation,50 W_regularizer=l1l2(self.l1_penalty, self.l2_penalty),51 init=self.init))52 model.add(Dropout(self.dropout_probability))5354 model.add(Dense(55 n_dims,56 activation=self.output_activation,57 W_regularizer=l1l2(self.l1_penalty, self.l2_penalty),58 init=self.init))5960 loss_function = make_reconstruction_loss(n_dims)6162 model.compile(optimizer=self.optimizer, loss=loss_function)63 return model6465 def fill(self, missing_mask):66 self.data[missing_mask] = -16768 def _create_missing_mask(self):69 if self.data.dtype != "f" and self.data.dtype != "d":70 self.data = self.data.astype(float)7172 return np.isnan(self.data)7374 def _train_epoch(self, model, missing_mask, batch_size):75 input_with_mask = np.hstack([self.data, missing_mask])76 n_samples = len(input_with_mask)77 n_batches = int(np.ceil(n_samples / batch_size))78 indices = np.arange(n_samples)79 np.random.shuffle(indices)80 X_shuffled = input_with_mask[indices]8182 for batch_idx in range(n_batches):83 batch_start = batch_idx * batch_size84 batch_end = (batch_idx + 1) * batch_size85 batch_data = X_shuffled[batch_start:batch_end, :]86 model.train_on_batch(batch_data, batch_data)87 return model.predict(input_with_mask)8889 def train(self, batch_size=256, train_epochs=100):90 missing_mask = self._create_missing_mask()91 self.fill(missing_mask)92 self.model = self._create_model()9394 observed_mask = ~missing_mask9596 for epoch in range(train_epochs):97 X_pred = self._train_epoch(self.model, missing_mask, batch_size)98 observed_mae = masked_mae(X_true=self.data,99 X_pred=X_pred,100 mask=observed_mask)101 if epoch % 50 == 0:102 print("observed mae:", observed_mae)103104 old_weight = (1.0 - self.recurrent_weight)105 self.data[missing_mask] *= old_weight106 pred_missing = X_pred[missing_mask]107 self.data[missing_mask] += self.recurrent_weight * pred_missing108 return self.data.copy()Whew, that was a large chunk of code. The important part is updating our data where values are missing. We use some predefined weight along with the predictions of our NN to update only the missing value cells. Here is a diagram of our model:
3. Evaluation
Let’s see how well our Autoencoder can impute missing data, shall we?
3.1 Preparing the data
We will use the encoding technique for our categorical data as discussed above. Let’s load (and drop a column with missing values from) our dataset.
1df = pd.read_csv("data/mushrooms.csv")2df = df.drop(['sroot'], axis=1)3.1.1 Making data dissapear
We will use MCAR as a driving process behind making missing data. Each data point in our dataset has 0.1 probability of being set to NaN.
1prob_missing = 0.12df_incomplete = df.copy()3ix = [(row, col) for row in range(df.shape[0]) for col in range(df.shape[1])]4for row, col in random.sample(ix, int(round(prob_missing * len(ix)))):5 df_incomplete.iat[row, col] = np.nan3.1.2 Encoding
Let’s one-hot encode our dataset using panda’s get_dummies and apply our missing data points accordingly:
1missing_encoded = pd.get_dummies(df_incomplete)23for col in df.columns:4 missing_cols = missing_encoded.columns.str.startswith(str(col) + "_")5 missing_encoded.loc[df_incomplete[col].isnull(), missing_cols] = np.nan3.2 Working out our Encoder (or just training it)
Finally, it is time to use our skillfully crafted model. We will train it for 300 epochs with a batch size of 256. All else is left to it’s default options.
1imputer = Autoencoder(missing_encoded.values)2complete_encoded = imputer.train(train_epochs=300, batch_size=256)1observed mae: 0.2307377938272observed mae: 0.03718809099973observed mae: 0.02687123573694observed mae: 0.02323425791295observed mae: 0.02107639753676observed mae: 0.0191748421237That’s all folks! Just kidding. The error seems to decrease so it might be wise to train for a few (or much more) epochs.
That run surprisingly fast on my 2.6 Ghz i7 (2013) with 16 Gb DDR3 ram. Should definitely try it out on a CUDA-enabled machine.
3.3 How well we did?
We have to do a little more housekeeping before answering that one. Here is an example of a row from our imputed dataset:
1complete_encoded[10, :10]1array([ 1.00000000e+00, 0.00000000e+00, 0.00000000e+00,2 0.00000000e+00, 0.00000000e+00, 0.00000000e+00,3 0.00000000e+00, 1.00000000e+00, 4.18084731e-17,4 5.83600606e-20])Not what you expected? That’s all right. Let’s use maximum likelihood estimation (MLE) to pick winning prediction for each missing data point.
1def mle(row):2 res = np.zeros(row.shape[0])3 res[np.argmax(row)] = 14 return res56col_classes = [len(df[c].unique()) for c in df.columns]78dummy_df = pd.get_dummies(df)910mle_complete = None1112for i, cnt in enumerate(col_classes):13 start_idx = int(sum(col_classes[0:i]))14 col_true = dummy_df.values[:, start_idx:start_idx+cnt]15 col_completed = complete_encoded[:, start_idx:start_idx+cnt]16 mle_completed = np.apply_along_axis(mle, axis=1, arr=col_completed)17 if mle_complete is None:18 mle_complete = mle_completed19 else:20 mle_complete = np.hstack([mle_complete, mle_completed])Let’s see what we’ve got:
1mle_complete[10, :10]1array([ 1., 0., 0., 0., 0., 0., 0., 1., 0., 0.])That’s more like it! Now we just have to reverse that get_dummies encoding…
1def reverse_dummy(df_dummies):2 pos = defaultdict(list)3 vals = defaultdict(list)45 for i, c in enumerate(df_dummies.columns):6 if "_" in c:7 k, v = c.split("_", 1)8 pos[k].append(i)9 vals[k].append(v)10 else:11 pos["_"].append(i)1213 df = pd.DataFrame({k: pd.Categorical.from_codes(14 np.argmax(df_dummies.iloc[:, pos[k]].values, axis=1),15 vals[k])16 for k in vals})1718 df[df_dummies.columns[pos["_"]]] = df_dummies.iloc[:, pos["_"]]19 return df2021rev_df = reverse_dummy(pd.DataFrame(data=mle_complete, columns=dummy_df.columns))22rev_df = rev_df[list(df.columns)]How much incorrect data points we have?
1incorrect = (rev_df != df)2incorrect_cnts = incorrect.apply(pd.value_counts)3incorrect_sum = incorrect_cnts.sum(axis=1)4incorrect_sum[1]13807.0Whoops, that sounds like a lot. Maybe we didn’t do well overall? But how much are missing?
1missing = df_incomplete.apply(pd.isnull)2missing_cnts = missing.apply(pd.value_counts)3missing_sum = missing_cnts.sum(axis=1)4missing_sum[1]117873Whew, that is a lot, too! Okay, I’ll stop teasing you, here is the accuracy of our imputation:
1accuracy = 1.0 - (incorrect_sum[1] / missing_sum[1])2print(accuracy)10.786997146534Roughly 79%. Not bad considering the amount of data we have. But we haven’t compared that to other approaches for data imputation (well, I have).
4. Conclusion(s)
And once again, the day is saved thanks to the powerp… the Autoencoder. It was quite fun and easy to build this model using Keras. Probably it will be even easier in the near future since Keras will be integrated directly into TensorFlow. Some questions that require further investigation remain unanswered:
- Is this model effective for categorical data imputation?
- Was our data highly correlated and easy to predict?
- Can other models perform better? If so, when?
- Can we incorporate uncertainty into our imputation? So we have a degree of belief of how good the imputed value is.
You can think about those questions. I am gonna go eat some mushrooms.
References
Keras - The library we used to build the Autoencoder
fancyimpute - Most of the Autoencoder code is taken from this awesome library
Autoencoders - Unsupervised Feature Learning and Deep Learning on Autoencoders
Denoising Autoencoders - Tutorial on Denoising Autoencoders with short review on Autoencoders
Data Imputation on Electronic Health Records - Great use of Deep Autoencoders to impute medical records data
Data Imputation using Autoencoders in Biology - Another great use of Autoencoders for imputation
Share
Want to be a Machine Learning expert?
You'll never get spam from me